Volume: 13 Issue: 3
Isolation and Identification of Various Candida Species from Clinical Samples at a Tertiary Care Hospital of Southern Rajasthan
Year: 2025, Page: 139-142, Doi: https://doi.org/10.47799/pimr.1303.25.12
Received: May 31, 2025 Accepted: May 30, 2025 Published: Dec. 30, 2025
Abstract
Introduction: Candida species are yeast-like fungi that normally inhabit humans as harmless commensals but can become opportunistic pathogens when host resistance is compromised. Among the Candida species, Candida albicans is typically regarded as the main pathogen. Over the past few decades, there has been a noticeable rise in the prevalence of non-albicans species. Species-level identification of Candida spp. is of clinical significance due to the variability in virulence and antifungal resistance among them. Objective: The objective of this research is to isolate and identify the species of Candida from diverse clinical samples within a tertiary care hospital, to ascertain the range of Candida infections present in clinical specimens, and to characterize the Candida species that have been isolated. Methods: Candida isolates from various clinical samples like urine, sputum, tracheal swab, vaginal swab, and endotracheal secretions were included in this study. After initial identification of Candida species by conventional methods, Candida spp. were inoculated onto CHROM agar and incubated at 37°C for 48 to 72 hours, with species identification based on the type and colour of colonies on the CHROM agar medium. Results: In this present study, a total of 45 Candida species were isolated from various clinical specimens, 20 from urine, 12 from sputum, 6 from tracheal swab, 6 from vaginal swab and 1 from endotracheal secretions as shown in Table 2. Among 45 Candida isolates, 25 (55.56%) were of Candida albicans, 13 (28.89%) of Candida tropicalis, 6 (13.33%) of Candida galbrata and 1(2.22%) of Candida krusei. Conclusion: In addition to Candida albicans, non-albicans Candida species such as C. tropicalis, C. krusei, C. glabrata, and C. dubliniensis are being more frequently obtained from clinical specimens. Accurate species identification of Candida is crucial for effective treatment, as not all species respond uniformly to therapy, and there is a growing concern over antifungal resistance.
Keywords: Candida; Chrome agar; C. Tropicalis; C. Glabrata
INTRODUCTION
Candida species are yeast-like fungi that typically exist as harmless commensals on mucosal surfaces of the human body, particularly in the gastrointestinal and genitourinary tracts. However, under certain conditions such as immunosuppression, invasive procedures, or antibiotic use, they can become opportunistic pathogens, causing a spectrum of infections ranging from superficial candidiasis to invasive, life-threatening disease.1, 2, 3 The genus Candida comprises over 170 species, of which more than 17 are known to cause human infections.1 Candida albicans is traditionally the most common species implicated in candidiasis. However, in recent decades, there has been a notable shift toward non-albicans Candida (NAC) species, such as C. tropicalis, C. parapsilosis, C. glabrata, and C. krusei.4, 5, 6, 7 Among these, C. tropicalis is frequently isolated in clinical settings in India.1, 8 Risk factors for candidiasis include prolonged hospitalisation, the use of indwelling medical devices, immunosuppressive therapy (e.g., corticosteroids, chemotherapy), broad-spectrum antibiotic use, and HIV/AIDS.1, 9 C. krusei is intrinsically resistant to fluconazole, while C. glabrata exhibits reduced susceptibility. C. tropicalis is noted for its biofilm-forming ability on catheters and prosthetic devices, contributing to its resistance profile. Resistance to azoles, particularly fluconazole, has been increasingly reported in both C. albicans and C. tropicalis.1, 10 Species-level identification is crucial due to the varying antifungal resistance patterns and virulence among Candida species.11 Additionally, distinguishing between colonisation and true infection—especially in non-sterile specimens such as sputum or endotracheal secretions—is essential for accurate diagnosis and effective treatment. The present study aims to isolate and identify Candida species from a range of clinical specimens in a tertiary care hospital, assess their pathogenic potential, and correlate findings from sugar assimilation testing with CHROMagar identification.
MATERIALS AND METHODS
Study Design and Setting
A retrospective study was conducted in the Department of Microbiology, GBH Hospital, Udaipur, from 16th September 2024 to 16th November 2024, after approval from the Institutional Ethics Committee [AIIMSUDR/2025/6392].
Sample Collection: In the present study, a total of 45 Candida isolates were recovered from various clinical specimens. The majority were obtained from urine samples (n=20), followed by sputum (n=12), tracheal swabs (n=6), vaginal swabs (n=6), and endotracheal secretions (n=1)
To address the concern regarding sample quality: Bartlett’s grading was performed for sputum samples to ensure specimen adequacy. Endotracheal secretions were evaluated alongside clinical signs, duration of intubation, and radiological findings to differentiate colonization from infection. Although tracheal swabs are not routinely recommended, they were included in this retrospective analysis and are reported with limitations.
Isolation and Culture: Clinical specimens were first examined by Gram stain. Samples were inoculated onto Sabouraud Dextrose Agar (SDA) and incubated at 37°C for 48–72 hours, following standard guidelines for fungal culture to ensure adequate recovery of yeast colonies.
Identification of Candida Species
Germ tube test was conducted on all isolates. Germ tube–positive isolates were presumptively identified as C. albicans or C. dubliniensis.
Cornmeal Agar (CMA) with Tween 80 was used to observe chlamydospores formation, hyphae/Pseudohyphae and colonie morphology, which aided in species-level differentiation. Growth at 45°C was used to differentiate C. albicans (positive) from C. dubliniensis (negative).
Pathogenic vs. Commensal Assessment
The clinical importance of each isolate was evaluated by correlating:
-
The site of isolation
-
Clinical diagnoses and symptom presentations
-
Pertinent laboratory parameters (e.g., pyuria in urine specimens)
-
The application of invasive devices (e.g., Foley catheters, central lines)This helped distinguish pathogenic isolates from colonizers or commensals.
Statistical Analysis
Descriptive statistics were used to calculate frequencies and percentages of each Candida species by specimen type. Species distribution and association with sample types were evaluated. The correlation between sugar assimilation tests and CHROMagar colony morphology was analysed for accuracy. All data were compiled in Microsoft Excel and analysed using SPSS version 26.0.
Table 1: Colour characteristics of various Candida species on CHROM agar
|
Name of Candida species |
Colour on CHROM agar |
|
Candida albicans
|
Light green |
|
Candida tropicalis
|
Metallic blue |
|
Candida krusei
|
Rose pink |
|
Candida glabrata
|
White |
|
Candida parapsilosis
|
Pale cream |
|
Candida dubliniensis
|
Dark green |
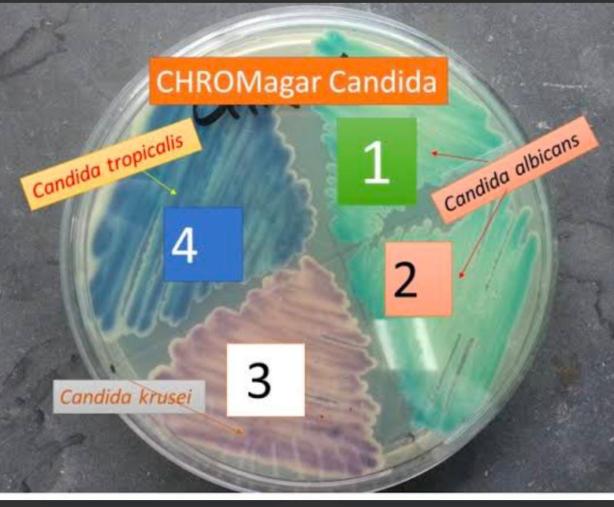
Figure 1: CHROM agar showing growth of different colonies of Candida species
RESULTS
C. albicans remained the predominant species, but a significant proportion (44.44%) of isolates were non-albicans Candida species, reflecting an emerging epidemiological trend. The distribution of isolates was as follows: 20 from urine, 12 from sputum, 6 from tracheal swabs, 6 from vaginal swabs, and 1 from endotracheal secretions, as shown in Table 2. All specimens were incubated for 48–72 hours at 37°C as per the recommended guidelines for Candida isolation.” Among the 45 Candida isolates, 25 (55.56%) were identified as Candida albicans, 13 (28.89%) as Candida tropicalis, 6 (13.33%) as Candida glabrata, and 1 (2.22%) as Candida krusei, as shown in Table 3. Gender-wise distribution revealed a higher prevalence in females (n=32) compared to males (n=13). Quality control of sputum specimens was done using Bartlett’s criteria. Isolates from endotracheal secretions and tracheal swabs were interpreted cautiously, with clinical correlation and available patient data considered to differentiate colonisation from infection.
Species identification was primarily performed using CHROMagar Candida. However, confirmatory identification by sugar assimilation test was done for representative isolates and correlated well with CHROMagar results, showing good concordance.
Table 2: Number of Candida isolates from various clinical specimens
|
Clinical Specimen |
Total Candida Isolates (n=45) |
|---|---|
|
Urine |
20 |
|
Sputum |
12 |
|
Tracheal Swab |
6 |
|
Vaginal Swab |
6 |
|
Endotracheal Secretions |
1 |
Table 3: Distribution of different Candida species and gender-wise distribution
|
Candida Species |
No. of Isolates (%) |
Male |
Female |
|---|---|---|---|
|
Candida albicans
|
25 (55.56%) |
7 |
18 |
|
Candida tropicalis
|
13 (28.89%) |
3 |
10 |
|
Candida glabrata
|
6 (13.33%) |
2 |
4 |
|
Candida krusei
|
1 (2.22%) |
1 |
0 |
|
Total |
45 |
13 |
32 |
DISCUSSION
The present study confirms the predominance of Candida albicans among clinical isolates, although a significant proportion (44.44%) consisted of non-albicans Candida (NAC) species, including C. tropicalis, C. glabrata, and C. krusei. This pattern aligns with several recent studies indicating an ongoing epidemiological shift.
A study by Kaup et al.12 reported 50% NAC isolates, with C. tropicalis accounting for 27.08%, closely matching our finding of 28.89%. Previous research by Manjunath et al. (2012) 13, Baradkar et al. (2010) 14, and Usharani et al. (2011) 15 also supports our observation that C. albicans remains the most frequently isolated species. However, reports by Istalingam et al.4 and Jayalakshmi L et al.16 demonstrated higher NAC isolation rates (56% and 68.57%, respectively), suggesting regional variation and an increasing prevalence of NAC species.
The presence of Candida in specimens such as sputum and endotracheal secretions must be interpreted cautiously. In our study, specimen quality was ensured using Bartlett’s scoring for sputum samples, and clinical history, radiological findings, and immune status were considered for invasive samples to distinguish colonisers from true pathogens.
CHROMagar Candida proved to be a useful preliminary tool for rapid species identification. However, the absence of Cornmeal Agar (CMA) use is a limitation. Sugar assimilation tests were performed on a subset of isolates and showed concordance with CHROMagar-based identification, supporting its reliability in our setting.
CONCLUSION
This study highlights a growing trend of non-albicans Candida species in clinical infections, although Candida albicans remains predominant. Rapid presumptive identification using CHROMagar Candida provides a practical approach, particularly in resource-limited settings. However, incorporating confirmatory methods such as sugar assimilation tests or CMA would enhance diagnostic accuracy.
Accurate species-level identification is essential for guiding antifungal therapy, especially given that some NAC species exhibit intrinsic resistance to commonly used antifungal agents. Furthermore, clinical correlation and appropriate specimen validation are crucial to distinguish colonisation from true infection, particularly in respiratory and urinary samples.
Future studies should include a larger sample size, comprehensive statistical analysis, and clinical outcome data to better understand the pathogenic role of various Candida species and their antifungal susceptibility patterns.
References
-
Kaur, Pavneet, Rawat, Tania, Sharma, Sarabjeet & Kaur, Parmeet . 2021. Speciation of candida species isolated in clinical samples in a tertiary health care centre in Northern India. IP International Journal of Medical Microbiology and Tropical Diseases 7(4):262–268.
-
Kanna, B Vignesh, G Amar Kumar, M Swapna & Easow, Joshy M . 2017. Isolation and identification of candida species from various clinical samples in a tertiary care hospital. International Journal of Research in Medical Sciences 5(8):3520–3522.
-
Anaissie, E J, McGinnis, M R & Pfaller, M A . 2009. Clinical Mycology (2). (p. 688 pages) Philadelphia: Churchill Livingstone
-
I Istalingam, D Vedachalam & S Rathinam . 2021. Isolation and speciation of candida from various clinical samples by using conventional method and chrom agar in a tertiary care centre. MedPulse International Journal of Microbiology 20(1):06–10.
-
Azad, M, Das, S & Kumar, A . 2019. Isolation, identification and anti-fungal susceptibility testing of candida species from various clinical specimens of ICU patients in a tertiary care hospital of Bhagalpur, Bihar. International Journal of Research and Review 6(9):115–122.
-
Mokaddas, Eiman M, Al-Sweih, Noura A & Khan, Zia U . 2007. Species distribution and antifungal susceptibility of Candida bloodstream isolates in Kuwait: a 10-year study. Journal of Medical Microbiology 56(2):255–259.
-
Srinivasan, L & Kenneth, J . 2006. Antibiotic Susceptibility of Candida Isolates in a tertiary care hospital in Southern India. Indian Journal of Medical Microbiology 24(1):80–81.
-
Mokaddas, Eiman M, Al-Sweih, Noura A & Khan, Zia U . 2007. Species distribution and antifungal susceptibility of Candida bloodstream isolates in Kuwait: a 10-year study. Journal of Medical Microbiology 56(2):255–259.
-
Jaggi, T, Urhekar, A D, Pai, C, Hodiwal, B A, Gore, S & Kar, H . 2014. Study of Candida species in various clinical samples in a tertiary care hospital. DHR International Journal of Medical Sciences 5(2):83–88.
-
Samyukthaa, A & Saikumar, C . 2017. Isolation, Identification and Speciation of Candida Species from Various Clinical Specimens in a Tertiary Care Hospital in Chennai. Scholars Journal of Applied Medical Sciences (SJAMS) 5(8F):3460–3468.
-
Offianan, Andre Toure, Ira, Ama Bonouma, Angora, Etienne, Vanga, Bosson Henriette, Sylla, Kady, Ako, Ako Berenger & Soumahoro, Adama . 2016. Species identification of Candida isolates in various clinical specimens and their antifungal susceptibility patterns in Cte dIvoire. African Journal of Microbiology Research 10(2):66–72.
-
Kaup, Soumya, Sankarankutty, Jaya, Balasubrahmanya, H V, Kulkarni, Suma & Nirmala, M . 2016. Speciation of Candida using HiCrome Candida Differential Agar. International Journal of Current Microbiology and Applied Sciences 5(7):267–274.
-
Manjunath, V, Vidya, G S, Sharma, A, Prakash, M R & Murugesh, . 2012. Speciation of Candida by Hicrome agar and sugar assimilation test in both HIV infected and non infected patients. International Journal of Biological & Medical Research 3(2):1778–1782.
-
Baradkar, V P, Mathur, M & Kumar, S . 2010. Hichrom candida agar for identification of candida species. Indian Journal of Pathology and Microbiology 53(1):93–95.
-
Usharani, A, Bharathi, M & Sandhya, C . 2011. Isolation and characterisation of candida species from oropharyngeal secretions of HIV positive individuals. Our Dermatology Online 2(3):119–124.
-
Jayalakshmi, L, Kumari, G Ratna & Samson, S H . 2014. Isolation, Speciation and Antifungal Susceptibility Testing of Candida from Clinical Specimens at a Tertiary Care Hospital. Scholars Journal of Applied Medical Sciences (SJAMS) 2(6E):3193–3198.
Copyright
©2025 (Jain) et al. This is an open-access journal, and articles are distributed under the terms of the Creative Commons Attribution License CC-BY 4.0. (https://creativecommons.org/licenses/by/4.0/) which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
Cite this article
Jain S, Kalra D, Mishra S. Isolation and Identification of Various Candida Species from Clinical Samples at a Tertiary Care Hospital of Southern Rajasthan. Perspectives in Medical Research. 2025;13(3):139-142 DOI: 10.47799/pimr.1303.25.12